2015 Library News Blog
Table of Contents:
Registration and access tips for BoardVitals (December 23, 2015) Mittens, hats, and scarves for The Mitten Tree (December 14, 2015) Bioinformatics Bites: MedGen (December 11, 2015)
Bioinformatics Bites: GEO2R (December 4, 2015)
New Resource: Quetzal Advanced Version (December 4, 2015)
Health Sciences Library: Web Satisfaction Survey Results (November 24, 2015) UpToDate app re-verification extended to 90 days! (November 24, 2015) Fitzsimons (November 18, 2015)
New e-books (November 11, 2015)
Bioinformatics Bites: Expression of a single gene in GEO Profiles (November 6, 2015) Art from CU Denver|Anschutz Medical Campus Community 2016 (November 5, 2015) Bioinformatics Bites: the GEO Databases (October 30, 2015)
New exam prep tool: BoardVitals (October 27, 2015) Library Access 24/7 (October 22, 2015)
Try out OvidToday: an app for browsing Ovid journals (October 16, 2015) Keep Calm and Embrace Conflict (October 13, 2015)
FYI: 2015 Legislative Blue Book (October 6, 2015)
Exciting News for Anschutz Medical Campus Faculty and Staff! FREE INTERLIBRARY LOAN (ILL)! (September 30, 2015)
Table of Contents (continued):
Clinical Corner: IOM’s “Improving Diagnosis in Health Care” (September 29, 2015) Bioinformatics Bites: Repurposing publicly available data (September 25, 2015) Clinical Corner: Acute Care for Older Adults (September 23, 2015)
Bioinformatics Bites: PubChem Bioassay (September 18, 2015) New e-books (September 17, 2015)
What are your three library wishes? (September 15, 2015)
Bioinformatics Bites: Creating a custom database to search with BLAST (September 11, 2015) Call for submissions – Art from the CU Denver | Anschutz Medical Campus Community 2016
(September 8, 2015)
Bioinformatics Bites: PubChem (September 4, 2015)
Private: Online Service Interruption: Labor Day, Monday, Sept 7, 6am-8am (September 4, 2015) The Library Has Gone Green! (September 3, 2015)
Private: Test out the library’s new responsive website (September 1, 2015) NCBI cracks 200 annotated eukaryotic genomes (September 1, 2015) Clinical Corner: Inpatient Glycemic Control (August 28, 2015)
Bioinformatics Bites: Primer BLAST (August 21, 2015) New ProQuest Interface (August 18, 2015)
Clinical Corner: Seizure/Epilepsy Videos (August 17, 2015)
Bioinformatics bites: How do I find primer binding sites? (August 14, 2015) Private: Off-Campus Access is changing! (August 14, 2015)
Table of Contents (continued):
New article linker enhancement: Sidebar Helper (August 10, 2015) Bioinformatics Bites: R tutorials for beginners (August 7, 2015) Clinical Corner: High-Value, Cost-Conscious Care (August 3, 2015) Bioinformatics bites: Navigating BLAST results (July 31, 2015) NCBI is innovating: give them your feedback (July 30, 2015)
Librarians in the Lab: Gates Biomanufacturing Facility (July 24, 2015) Bioinformatics bites: Constructing a BLAST query (July 24, 2015) Want to publish in Nature? (July 20, 2015)
Clinical Corner: Joanna Briggs Institute database (July 15, 2015)
Critical need for platelets — Children’s Hospital has 3x number of Hematology/Oncology patients (July 10, 2015)
Search box on library’s website will be in header of every page (July 6, 2015) Bioinformatics Bite: How to find the transcription start site of a gene. (July 2, 2015) Bioinformatics Bite: Searching the gene database (June 26, 2015)
New Exhibit – Michael Keyes: Stories & Seasons in Woodcut (June 23, 2015) Bioinformatics bite: How to find the official name of a gene (June 19, 2015) Cochrane Review Matchmaking Through Social Media (June 17, 2015)
SciENcv — Converting Profiles that Use the Old NIH Biosketch Format to the New NIH Biosketch Format (June 17, 2015)
Table of Contents (continued):
Announcing Bioinformatics Bites (June 12, 2015) New Staff at HSL – Christi Piper (June 11, 2015)
Leaving Anschutz Medical Campus? Suggestions for a smooth transition (June 9, 2015) New Staff at HSL – Kristen Desanto (June 8, 2015)
New e-books (June 4, 2015)
PubVenn: Visualize your PubMed searches (June 3, 2015)
“Chasing the Cure” – Colorado Springs and Tuberculosis (May 31, 2015) Great Moments in Medicine and Great Moments in Pharmacy (May 28, 2015) Special Collections Featured Book for April (May 6, 2015)
Upcoming Strauss-Wisneski Lecture on Integrative Medicine (April 28, 2015) Random from the Repository (April 23, 2015)
Rare Book Profile: Dell’anatomia, a facsimile of Leonardo da Vinci’s notebook. (April 23, 2015) New e-books (April 23, 2015)
New exhibit combines research and fashion (April 6, 2015) The Fondest of Farewells (March 30, 2015)
SAS now available at the HSL (March 27, 2015)
Article Linker problem from PubMed (March 20, 2015) New Reading Room Exhibit Case (March 18, 2015)
Open Access Fund for Spring 2015: Now Closed (March 17, 2015) Informatics Workshop Series (March 16, 2015)
Table of Contents (continued):
The Writing Center: What it can do for you (March 13, 2015) Tower Room updated with permanent PC (March 12, 2015) New Additions to Amesse (March 10, 2015)
Reference Collection Overhaul (March 9, 2015) Upcoming Strauss-Wisneski Lectures (March 9, 2015) STAT!Ref’s Bookshelf (March 6, 2015)
Webinar March 5, 2015: NCBI and the NIH Public Access Policy: PMC Submissions, My NCBI, My Bibliography and SciENcv (March 2, 2015)
Rare Book Profile: Thomas Willis’ Cerebri Anatome cue Accessit Nervorum Descriptio et Usus. (February 27, 2015)
NIH Workshop on Building a Precision Medicine Research Cohort (February 11, 2015) New DynaMed App! (February 6, 2015)
Mobile whiteboards: Round and round they go (February 6, 2015) New e-books in R2 (January 30, 2015)
Updated: Personal account access problem in Ovid (January 29, 2015) 2015 Annual MSA Capstone Event: Call for Judges (January 29, 2015) HSL Informatics Workshop Series (January 27, 2015)
Blood Drive for Liam on Friday, Jan 30th! (January 27, 2015)
Earn while you learn: Student employment at the Health Sciences Library (January 22, 2015) Tax Time Tips (January 20, 2015)
Table of Contents (continued):
Portable physical therapy table now available for check out (CABI) (January 14, 2015) Colorado PROFILES Now Featuring Altmetrics (January 12, 2015)
New Resource: Joanna Briggs Institute EBP (JBI) (January 7, 2015)
Access Services Conference – November 2014 Georgia Tech University (January 7, 2015) Exhibit – Art from CU Denver | Anschutz Medical Campus (January 6, 2015)
Downloading E-books in ClinicalKey (January 5, 2015)
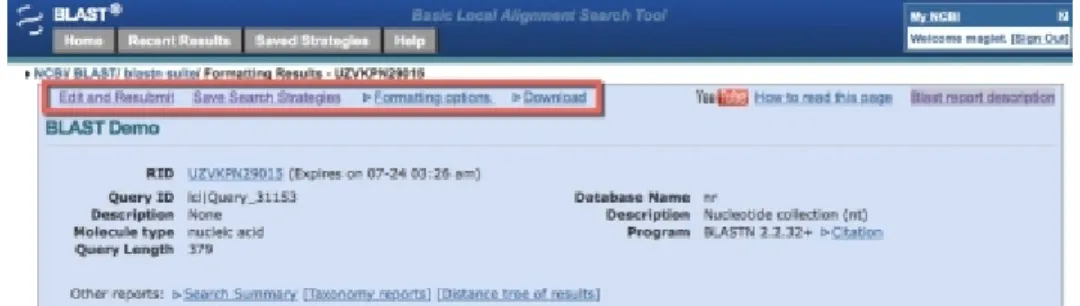
Registration and access tips
for BoardVitals
Users must register to access BoardVitals. When you first set up an account, you will only see this screen until you fully activate your account through the follow-up email:
After you click the link in the follow-up email, you should have full access to BoardVitals and the screen should look like this:
You can access BoardVitals on the library’s database page, or through STAT!Ref:
Question bank areas include: USMLE Step 1 USMLE Step 2 USMLE Step 3 Shelf Exams NBDE I NBDE II NAPLEX NCLEX RN PANCE
Faculty, program directors, etc. can be designated as administrators in order to track student/departmental performances and utilize the custom exams for students. Contact the library if you are interested in this feature or have any questions!
Mittens, hats, and scarves for The
Mitten Tree
The Health Sciences Library has put up our annual Mi en Tree on the first floor main entrance of the library. All donations will benefit The Comitis Crisis Center.
What can I donate? We invite everyone to
donate new or newly-made mi ens, hats, and scarves. We also welcome towels, twin sheets, and toiletries (toothbrushes, toothpaste, deodorant, soap, shampoo, etc.)
How do I donate? Simply bring in the items and
hang them on the tree at the library.
Until when can I donate? The tree will be up
through January 8, 2016.
Who will my donation benefit? After the holidays, library staff will take all of the
items to the nearby Comitis Crisis Center. Comitis is a non-profit agency underneath the umbrella of Mile High Behavioral Healthcare. The shelter serves runaway/homeless children, homeless families, and homeless Veterans—with a particular focus on
homeless female Veterans and their families.
Questions: For more information or questions, contact Library Administration at
303-724-2138.
This bioinformatics bite is going to be a li le but more clinically oriented:
A patient is presenting with excess blood clo ing, which she thinks might be related to “something that runs in her family”. How do I find known diseases and genes (if any) that are associated with that phenotype?
A good place to start to look for information about symptoms and diseases that are related to genetics is MedGen. This database organizes information related to human medical genetics, like symptoms (clinical features), related genes, diseases, or genomic loci.
A perfectly reasonable approach would be to type “clo ing” into the MedGen search box. Here’s what those results look like:
There are 94 results, the first of which is a clo ing disorder, but one that is associated with too li le clo ing rather than too much clo ing. If you scroll down, you see records that are not actually diseases:
To find out what type of record you’re looking at, look at the text after concept ID (blue boxes). The screen captures above show a Disease or Syndrome, a Finding, and a
Pharmacologic substance. Notice that the diseases has links to other databases (green circle) and the others do not.
So how do we specify that we’re looking for a patient symptom related to a genetic disease? Like all the other NCBI databases, MedGen has field tags.
Here are some useful ones:
Clinical Features: short stature[clinical features] – records for diseases that are
associated with short stature
Related Genes: LMNB1[gene] – diseases associated with this gene Disease name: achrondroplasia[title] – this disease
Chromosome: 6[chromosome]- diseases associated with alterations to chromosome
6
Also, if you look back at the first screen shot, you can see a link that says “See MedGen results with clo ing as a clinical feature (5)“. MedGen automatically sensed that
clo ing was a clinical feature, or symptom, and narrowed your results down for you. Now we’ve narrowed the MedGen results to those that have clo ing listed as a clinical feature. If you read the description, you see that Factor V deficiency is the only one associated with excess clo ing. The record also shows what gene is associated with this disorder (F5) and links to descriptions from other resources
like GeneReviews and OMIM, as well as Professional guidelines and Recent clinical studies.
Bioinformatics Bites: GEO2R
This blog series has covered how to use both GEO Datasets (which holds both curated and uncurated datasets) and GEO Profiles (which holds expression profiles for
individual genes from curated data sets).
But what if you want to see expression profiles of a gene from an uncharted dataset? That’s where GEO2R comes in. Once you’ve identified a dataset by searching GEO Datasets, you can start using GEO2R in 5 easy steps:
1. Pick your experiment 2. Define sample groups 3. Assign samples to groups 4. Perform the test
5. Interpret the results table
Pick your experiment:
You need an accession number to start using GEO 2R. Which one do you use? Let’s use this dataset from Toxoplasma gondii as an example.
This record contains accession numbers (boxed in red) for the series, samples, and platform. GEO2R is looking for the Series Accession number, GSE73177. Enter this number into the search field in GEO2R, or click the Analyze with GEO2R link (blue arrow):
Define sample groups:
This experiment is measuring expression levels in 3 groups (parent strain, knockout strain, and complemented knockout strain**). Thus, we need to create 3 sample groups. To do this, click “Define Groups” link (green circle above.) This action activates a
popup that allows you to enter free text to name the groups (red box, wt, ko, and comp in the following example)
Assign samples to groups:
Now you need to tell the program which samples belong to each group by selecting the samples that you want to put into a group, then clicking on the group you want to add them 2 in the Define Groups popup. In the example below, I have selected the
complemented knockout samples (highlighted yellow) and will click the “comp” group to add them. After they are added to a group, the corresponding colors change to that of the group and the group column is populated, as in the case of wt and ko in the example. Repeat this process for all of your groups.
Perform the test:
Get a list of the top 250 differentially expressed genes using the default se ings*, scroll down and click the Top 250 bu on under the GEO2R tab.
A table containing the top 250 differentially expressed probes from the platform that the probe ID, p-value, adjusted p-value, F statistic and probe sequence.
Interpret the results table:
Clicking on the probe ID will show you a graph of the gene expression among the groups you have specified. You can also click Sample Values to get the number values represented on this graph.
In this case, it looks like the gene that is probed by 55.m10280_at is highly expressed in the knockout relative to the wild type, but doesn’t revert to wild type levels in the complemented strain.
To determine the gene name of the probe used in this experiment, visit
the corresponding platform record for this series. (You can find this by searching the Series accession in GEO datasets, and using the platform filter.) Then, scroll to the bo om of the page to see the platform data table, which gives probe IDs, identifiers for genes in the toxoplasma genome database, annotation,chromosome location and a description of the gene function if available. Search for the probe id in the table to find the corresponding gene. In this case, it’s a thioredoxin domain-containing protein.
But what if your gene of interest is not in the top 250? You can use the Profile
Graph tab to search by probe id.
GEO2R also has basic QC tools. You can see the value distributions across samples to identify large scale problems in the dataset using the value distribution tab:
Finally, you can retrieve the R script for the analyses run in the R script tab. NCBI also has a comprehensive tutorial on this tool if you’re interested. Let me know if you have any questions!
C. Tobin Magle, PhD, Biomedical Science Research Support Specialist
* For novice users, the default se ings are a good place to start. The calculation uses
the limma package, and you can view and change the default se ings by clicking the Options tab.
New Resource: Quetzal
Advanced Version
Que al
The library now has access to the Advanced Version of Que al! Find it on the library’s database page, or follow the links/directions below.
Version descriptions:
Basic (Free) – Enhanced linguistic searching of PubMed documents to find relevant
results.
Advanced – Includes patent, full-text searching and AHRQ Treatment Protocols. Note: When creating a new individual account, users must be on campus, logged in with their PassportID after following the link below (if off campus), or connected by VPN.
For new users:
1. Go to Que al.
2. Scroll down to the bo om of the page. 3. You will see the following message:
If it doesn’t say “University of Colorado – Anschu Medical Campus,” you are not on campus and will need to go through your Proxy Server or VPN
4. Click the registration bu on at the bo om of the page:
5. Fill out the Registration Form. Our organization should be pre-selected for “University of Colorado – Anschu Medical Campus”. If not, please select that from the drop-down list. [Note: a separate registration is required for each
individual user since each user uses their personal space to save searches, get weekly alerts, save results with personal annotations, and participate in private journal clubs.]
6. Read the Terms of Use and check the box at the bo om of the Registration page. 7. Click the Continue bu on.
8. You should see a message saying “Your user registration was successful. You may Login to Que al® Search now.” [IMPORTANT NOTE: If you see instead a new form that says “Choose your Subscription type”, then STOP. You have not been correctly recognized as being from University of Colorado – Anschu Medical Campus. Be sure you are on site or logged in through your organization’s Proxy Server. Or, make sure you are signing up for Que al® Advanced, not
Professional.]
9. Happy Searching! You can follow along with the Guided Tour on your first visit. This will help you understand how to use Que al® effectively. You can also check out the information and 30” videos shown in the Que al® Quick Help section on the home page (after logging in).
10. Any questions or problems? Contact Quertle at info@quertle.com.
For users with an existing Que al® Basic account:
1. Go to Que al.
2. Login to your existing account.
3. In the upper right corner of the search area, click on “Your Que al”. 4. Click on the My Profile tab. In the Organization drop-down list, be sure
our subscribing organization name (University of Colorado – Anschu Medical Campus ) is chosen. This should be the same name you see on the Que al welcome page in the “Happy News” message at the bo om of the page. If it is not the same, please select the correct Organization from the list.
5. Click on the My Account tab. You should see a Happy News message. [NOTE: If you do not see the Happy News message, then STOP. You have not been correctly recognized as being from University of Colorado – Anschu Medical Campus. Be sure you are on campus or logged in through your organization’s Proxy Server.] 6. Click “Change” on the Your Version line.
7. Choose “Advanced” in the dialog that opens and then click Submit. You may need to log in again.
8. On your My Account page, the line for “Your Version” should now display Que al® Advanced.
9. Enjoy searching all the content Que al® has to offer and the full set of features and functions.
10. Any questions or problems? Contact Quertle at info@quertle.com.
Non-AMC campus members can still register for access to the Basic (free) version: h ps://www.que al-search.info/
Health Sciences Library: Web
Satisfaction Survey Results
Web Satisfaction Survey Results
Thank you to our users for responding so enthusiastically to our survey! We surveyed users from November 11-20, 2015 and were thrilled to receive a total of 227 responses!
Q1. How happy are you with the library’s website? 226 responded to Q1.
78% reported being either happy or very happy with the library’s website.
Q2: How can we improve your website experience? 93 wrote responses to open-ended Q2.
32% of wri en responses noted that there was no need for improvements to the
Respondents suggested many improvements to the website design, navigation, search, and access. The suggestions provide many excellent opportunities for the library to continue to improve our users’ web experience. We also received many suggestions that were not related to the website. These are just a few of the many comments we received:
you are doing great. I use pubmed from google scholar all the time and when it stopped working got excellent help from your staff. THANKS
I think that you are always improving the website and keeping up with the recent
developments and discoveries.Tobin Magle is amazing. Well done all! Just an suggestion, can the library also provide access to common analysis software ? Since some bioinformatics, ensembl and NCBI classes are offered it seems to me that having access to softwares through the library will boost the use of the website/library and provide a huge service to students, staff and faculty who do not know from where to obtain these.
Have mini-workshops available online on how to search websites for information more efficiently
The biggest challenge I have is ge ing back to the main library screen from FindIT. If I go there at all, I pre y much have to go back to my bookmark (and log back in) to get to other library resources. I am a remote user (student). The other suggestion is more options for learning to use tools such as Endnote for those of us who can’t be on-campus!
Oftentimes I feel very lost when trying to figure out if I have access to a journal, where I go to click and how I find relevant information to what I’m looking for.
you do a fantastic job with on-line services and responses to questions.
I love having access to online articles! The more that are available the happier I will be. More clarity on first page. Appreciate greater recent access made to research resource. Simplify. Home page is very busy.
All the links I need are right on the front page. Very easy to access!
And LOTS more! Read the full report with all user feedback: HSL-Website-Satisfaction-Survey-Results-Nov2015
Thank you to the Web Working Group, and especially Cathalina Fontenelle, for their dedication to improving our website!
Questions, please contact Vivienne Houghton, MLIS, Web Services Librarian, at 303-724-2178 or vivienne.houghton@ucdenver.edu
UpToDate app re-verification extended
to 90 days!
UpToDate has extended the re-verification period for their app from 30 days to 90 days.
Options for re-verification:
There are multiple ways you can re-verify your access to UpToDate:
1. Go to UpToDate.com and click the log-in link on the top right and then enter your user name and password (do this when connected to the hospital or campus
network)
-OR-2. Access UpToDate via an embedded link within your EHR system (EPIC)
-OR-3. Use the UpToDate Mobile App from the campus network (for example, from the UCDenver wireless network)
-OR-4. Are you away from campus? Go to Up to Date via the HS Library website, or, log in with the off-campus link: h p://proxy.hsl.ucdenver.edu/login?
url=h p://www.uptodate.com/online/content/search.do?
unid=^u&srcsys=UTD148363&eiv=2.1.0. Log in through EZproxy with your library credentials, then log in at UpToDate with your UpToDate user name and password.
Need the app? Get the app: h p://hslibrary.ucdenver.edu/uptodate-app
Benefits:
-Free UpToDate Mobile App for your iOS, Android™ or Windows 8 device -Direct access to UpToDate from any internet browser
-Earn CME/CE/CPD credits when you research clinical questions If you encounter any problems, let the library know!
And LOTS more! Read the full report with all user feedback: HSL-Website-Satisfaction-Survey-Results-Nov2015
Thank you to the Web Working Group, and especially Cathalina Fontenelle, for their dedication to improving our website!
Questions, please contact Vivienne Houghton, MLIS, Web Services Librarian, at 303-724-2178 or vivienne.houghton@ucdenver.edu
Fitzsimons
The Anschu Medical campus is located on the former Fi simons Army Base, closed in 1999. Fi simons was opened in 1918, and was named in honor of Lt. Thomas Fi simons, of the Army Medical Corps, who was the first US officer killed in the First World War. The hospital was opened to care for returning soldier who suffered from respiratory disease. The Base remained a key Army Medical Center until its closure and the iconic main hospital, known as Building 500, is still the center of the campus. Visit the Library’s second floor exhibit space to further explore the history of
–Paul Andrews
Head and Neck Pathology
Essentials of Environmental Health
Bioinformatics Bites: Expression of a
single gene in GEO Profiles
This weeks bioinformatics bite will answer a question from the end of last week’s post:
What is the expression of ITGA2 gene in prostate cancer cells?
I’d check GEO profiles, because it is a gene centric question. Let’s start by typing in prostate cancer in the GEO profiles search box.
Note the link back to the GEO data set that this gene profile was derived from (green circle). You can also see the platform and the specific probe that measures this gene (orange box). Finally, you can see a cartoon of the expression level between sample and control on the right side of the record.
To narrow down your search to a specific gene, use the filters on the left to select an organism (red box) and a Gene symbol (blue box, make sure to check that you’re using the correct gene symbol). Let’s look for the ITGA2 gene in human derived samples.
After applying the filters (blue and red boxes), you can see the search strategy in the Search details (orange box).
To get a close up of the expression graph, click the cartoon on the right (green circle).
At a glance, you can see that ITGA2 expression goes up when the microarray miR-205 is expressed (red bars). It also indicates how highly this gene is expressed relative to other genes from the same sample by percentile (blue squares). It also lists the expression values from each sample in a table below, along with its rank.
If you need more information about how the samples were prepared, you can click on the GSM number in the table. From there, you can access general information about the experiment by clicking on the Series ID (GSE) on any sample page, or the original GEO profile record.
But what do you do if the gene you’re a empting to access data in an uncurated data set? NCBI has a tool for that: GEO2R. We will discuss how to use this tool next time.
C. Tobin Magle, PhD, Biomedical Science Research Support Specialist
Art from CU Denver|Anschutz Medical
Campus Community 2016
On exhibit: November 4, 2015 to January 31, 2016 Location: Third Floor Gallery
An exhibit of artwork created by faculty, staff and students of the University of
Colorado Denver. There are many talented artists among the faculty, staff and students on our campuses. This juried exhibition is an opportunity for us to learn about our talented co-workers, teachers, and students. This exhibition is presented by the Exhibits Commi ee of the Health Sciences Library.
Opening reception: November 12, 2015 from 3:00 pm to 5:00 pm — Please join us! CU Art 2016 page: h p://hslibrary.ucdenver.edu/exhibits-2016-cu-art
Bioinformatics Bites: the
GEO Databases
Wow! It’s been a long time since i’ve been able to take the time to write a post. I apologize. Sorry for the hiatus. I have been traveling and playing catchup and a ending meetings post travel. I hope to get back to my weekly posts now.
This week’s bioinformatics bites addresses finding gene expression information using GEO profiles.
Background: The Gene Expression Omnibus (GEO) was created by the NCBI to store gene expression data from microarray experiments. Most of the content is still
microarrays, but some NGS data is also present (with the raw reads in the SRA
database). Additionally, it now contains other types of high-throughput genomics data, like CHiP-chip.
GEO exists as two separate databases:
GEO DataSets: original submi er-supplied records and curated data sets
GEO Profiles: Expression profiles of individual genes from curated GEO data sets GEO Data Sets, which are labeled with GDS numbers, have three major components submi ed:
Series (GSE) – List of expression profiles that conducted for the experiment (test,
control, replicates)
Samples (GDS) – Information about the biological samples used in the
experiments, including extraction procedures
Platforms (GPL) – What platform the samples were run on (like Affymetrix Mouse Genome 430 2.0 Array)
All 3 of these components are necessary to make use of the study. Both samples and series link to the CEL files, and platform gives you information about the chip the samples were run on. NCBI is working to assemble all of the components from each submi ed study into a curated DataSet, but there is some lag in the process. NGS studies can’t be curated at this time.
After curation, the data sets are broken out by gene instead of experiment and the data are loaded into GEO profiles.
GEO DataSets results
This looks a lot like the output of a Gene search, but with different filters. You can see in the red box that there are over 1000 cancer data sets in GEO, and that the top data set has 6 samples by following the red arrow. You can filter by study type (blue box), things like tissue or strain (purple box), or Organism (orange arrow). You also get a search details box, which shows that MeSH terms are applied to your text search, just like PubMed.
Now that we have a general idea of how the data bases are structures, I will answer a practical question in next week’s post:
What is the expression of ITGA2 gene in prostate cancer cells?
See you next week
New exam prep tool: BoardVitals
You can access BoardVitals on the library’s database page, or
through STAT!Ref:
Note: Users must register to access BoardVitals
Question bank areas include: USMLE Step 1 USMLE Step 2 USMLE Step 3 Shelf Exams NBDE I NBDE II NAPLEX NCLEX RN PANCE
Faculty, program directors, etc. can be designated as administrators in order to track student/departmental performances and utilize the custom exams for students. Contact the library if you are interested in this feature or have any questions!
Library Access 24/7
The Library is open 24/7 to provide additional study/research space for students, staff and faculty of CU. Security staff will do occasional walkthroughs of the building during unstaffed hours. If you need headphones, markers, or a print job under $5, those items will be available at the service desk on an honor policy. Please return items when finished.
Don’t forget to take regular breaks from your studies during study sessions. The library has a coffee machine located on the south side of the building on the 1st floor near the restrooms. We also have a vending area on the north end of the building on the 1st floor, for snacks and soda. Fell free to bring your own snacks and use in the fridges on all floors. Soon there will be two Keurig machines to make coffee on 2 and 3 floor. Water bo le stations are located next to the restrooms on each floor on the south side of the building.
If you have any questions or comments about 24/7 please contact library staff by emailing circ.library@ucdenver.edu , ask staff at service desk, or give us a call at 303-724-2152 for phone questions. For more information visit our website site page @ h p://hsl.ucdenver.edu/about/hours with more info about 24/7 & Badge Access.
Try out OvidToday: an app for browsing
Ovid journals
Want a way to keep current with Ovid journals? Try OvidToday
Benefits:
Browse specialty categories and select journals to follow Stay current with the latest journal content
Read the PDF of any article
Add articles to a personal reading list for later use, even offline Access journals at home or work with remote authentication Directions:
1) Download the free OvidToday app
Available for free. Get it on Google Play (play.google.com) or iTunes (itunes.apple.com)
2) Tap “Let’s get started” and follow the on-screen instructions
If you are on campus/logged in on the network, the app should automatically recognize you. Off campus? First log in to the library’s Ovid link or go through VPN so the system recognizes you as an affiliated user.
3) Enter an Ovid Personal Account ID
If you don’t have a Personal Account, OvidToday will prompt you to create one and provide steps.
4) Start browsing favorite journals
You will have access to the most recent issues of the library’s Ovid-subscribed journal content; new issues are added automatically once available on Ovid.
More information…
Keep Calm and Embrace Conflict
All are invited to a special presentation from Joseph P. Folger (Temple University) that will discuss how we can do be er to address and respond to conflicts encountered in our work and personal lives.
Date: October 14, 2015 Time: Noon – 1 pm Locations:
CU Anschu : ED 2 North, Room 1102 CU Denver: Auraria Library
Lunch will be provided! No RSVP needed! This event is co-sponsored by:
Health Sciences Library, Facilities Management, Ombuds Office, Human Resources / Campus Life Commi ee, Office of Academic Resources and Services (OARS) and Auraria Library
FYI: 2015 Legislative Blue Book
The Blue Book provides voter information on ballot measures.The Colorado 2014 Legislative Blue Book is available in pdf and mp3 formats (courtesy of the Colorado Talking Book Library).
The purpose of the ballot information booklet is to provide voters with the text, title, and a fair and impartial analysis of each initiated or referred constitutional amendment, law, or question on the ballot. The analysis must include a summary of the measure, the major arguments both for and against the measure, and a brief fiscal assessment of the measure. The analysis may also include any other information that will help voters understand the purpose and effect of a measure.
Article V, Section 1 (7.5), Colorado Constitution, and Section 1-40-124.5, Colorado Revised Statutes, require the Legislative Council Staff to prepare the ballot information booklet prior to each election in which a statewide issue will appear on the ballot.
Need to register to vote? If you wish to receive a mail-in ballot register as soon as possible! If you need information about voting in local elections contact your city or county clerk. Happy voting!
[Amanda Langdon]
Exciting News for Anschutz Medical
Campus Faculty and Staff! FREE
INTERLIBRARY LOAN (ILL)!
Beginning July 1, 2015, most CU-Anschutz Medical Campus Faculty and Staff will now receive FREE INTERLIBRARY LOAN (ILL)! This includes paid University faculty and staff of CU Anschutz, faculty paid by affiliates, volunteer clinical/faculty, retired CU Anschutz faculty, CU Anschutz fellows, CU Anschutz visiting scholars and students, and UPI employees. CU
Anschutz students and residents will continue to receive no-cost ILL as well.
Some affiliated users will still be charged services fees ($8/filled request, $18/rush). This includes CU Anschutz alumni who live in Colorado, CU Anschutz volunteer staff, UCH paid staff, UCH volunteers, other CU faculty, staff, and students, CCML members, and residents paid by other health organizations.
There will be no changes in current fees for all other users ($18/filled request, $48/rush). This includes Colorado public, non-university fellows, non-university visiting scholars, and all others not listed above.
Please be advised that this is a 2-year pilot program, 2015-2017.
Sign up for an ILLiad account today!
If you have any questions about your affiliation and eligibility, please contact the ILL office at 303-724-2111 or copydocs@ucdenver.edu with any and all questions.
[Brittany Heer, Library Technician II, Interlibrary Loan]
Clinical Corner: IOM’s “Improving
Diagnosis in Health Care”
On September 22, 2015, the Institute of Medicine (IOM) released a new report titled “Improving Diagnosis in Health Care.” This continues the series of reports that began in 1999, the first being “To Err is Human: Building a Safer Health System.”
The National Academies Press is currently offering a prepublication print copy for sale, or you can pre-order a paperback. But there are also several free options: you can read it online in HTML format, or download a copy in PDF format. To download a PDF you can either create a free MyNAP account or download it as a guest.
To access the report, go to the following
link: http://iom.nationalacademies.org/reports/2015/improving-diagnosis-in-healthcare
(From this page, you can also access a brief report, list of recommendations, and resources for improving communication in the REPORT AT A GLANCEbox.)
Click Download the report for free.
A new page will open. From there, select either Download Free PDF or Read Online (on the right-hand side of the page).
Kristen DeSanto, MSLS, MS, RD, AHIP Clinical Librarian
kristen.desanto@ucdenver.edu • 303-724-2121
Bioinformatics Bites: Repurposing
publicly available data
I’m going in a bit of a different direction with this bioinformatics bite segment. Instead of explicitly describing how to use a database or tool, I wanted to tell you all about a little project that I’m working on with a vet student from CSU.
Just because I don’t have a lab or large amounts of research funding doesn’t mean I can’t do science! There’s a wealth of bioinformatic data publicly available online and some user friendly tools that are available. We’re using these freely available resources to ask questions about how
the distribution of microbes in the environment correlates with specific landmarks.
Our research question is as follows: Are there differences in microbial populations at sites that are close to zoos in the NYC area compared to the farther away. To address this question, we are using data from the PathoMap project, which swabbed surfaces in transit stops all over New York City. (This project is also being expanded to the top 10 cities worldwide for public transit ridership by a project called MetaSub.) See this publication for more information.
Pathomap main page
We determined test and control sites by looking up how city planners determine the distance people are willing to walk to a transit stop. We are also using a tool called GeneGis 2 to analyze and visualize these data. See this publication for more information on GenGIS.
GenGIS output from their wiki
Repurposing data forces you to think differently. Our research question was designed in the context of what data and tools were publicly available. This type of research will only get easier as the mindset of creating well-designed community resources expands. Initiatives like Big Data to Knowledge (BD2K) and the Center for Open Science are driving this new trend.
If you’re curious about data repurposing, or how to find datasets and tools, please see
the bioinformatics section of our new Research Support Pages or set up a consultation to discuss your questions.
Clinical Corner: Acute Care for
Older Adults
Today’s Department of Medicine Grand Rounds featured an informative and entertaining presentation by Dr. Samir Sinha, from Toronto’s Mount Sinai and University Health Network Hospitals. Dr. Sinha, whose presentation was titled “Rethinking Traditional Models of Acute Care for Older Adults,” discussed how current healthcare delivery in Canada and the United States can negatively impact older adults. He presented the Acute Care for Elders (ACE) Strategy, employed at his hospital, as an alternative.
The following articles and resources, many of which were discussed by Dr. Sinha, are related to the topic of patient and system outcomes for older adults:
Bioinformatics Bites:
PubChem Bioassay
The NCBI insights blog has another wonderful post about PubChem.
h p://ncbiinsights.ncbi.nlm.nih.gov/2015/09/11/finding-chemical-probes-and-modulators-the-hunt-for-new-chemical-reagents-and-medicines/
This week, the post focuses on using the Bioassay database and how it can help identify chemical reagents and drugs.
Tobin Magle, PhD, Biomedical Sciences Research Support Specialist
New e-books
You can find these e-books on our e-books page or in the catalog, or follow the links below:
Handbook of Cognitive–behavioral Therapies
Advanced practice nursing ethics in chronic disease self-management
Emergency Public Health : Preparedness and Response
What are your three library wishes?
If the Library Genie granted you three library wishes, what would they be? Now you have the opportunity to let us know!
Submit your three wishes to the library genie today! Your wishes will be
anonymous, but if you’d be willing to talk more with us about your wishes you can include your name and e-mail address. Are there resources or services you’d like to see the library offer?
Has the library implemented enhancements that you’d like to see more of? How could the library be er assist you with your research, education or clinical needs?
Let us know – the Library Genie is accepting wishes through September 30th!
Bioinformatics Bites: Creating a custom
database to search with BLAST
This week’s Bioinformatics Bite will show you how to run a BLAST search in
a custom database that you create using Entrez queries. Also, if you’re interested, NCBI has a YouTube video that covers this topic.
To review, there are 2 ways to search the NCBI databases:
1. Sequence homology via BLAST
2. Text via Entrez
Remember Entrez? This search engine allows you to apply field tags to refine your search results. These field tags vary by database, which makes sense because different
datatypes necessitate different contextual fields. For example, you can find the field
tags for the sequence databases (Nucleotide, Protein, GSS, EST) here.
Let’s go back to our Constructing a BLAST Query post from last month. We’re going to focus on
Step 4: Choose your database (search set)
Here are my parameters:
1. Algorithm type: nucleotide BLAST
2. Query: NM_001182936.1(Saccharomyces cerevisiae S288c Ras family GTPase RAS2) 3. Search name: yeast ras2
4. Database: refseq_RNA
5. Specific algorithm: blastn (somewhat similar sequences)
I’m starting with a pre-made database and we’ll refine from there. Enter this information into nucleotide BLAST query page and hit BLAST.
Here is a graphic summary of the results:
(click to enlarge)
This action will open a new window.
The results are from 3 taxonomic groups: yeast, animals and protists. Let’s zero in on the animal group.
First, return to the original BLAST page.
Then, change the view so we only see sequences from animals by clicking Forma ing
options above the search summary and typing animals in the Organism field in
the Limit results section. The taxid for animals will autofill as you type.
The top result from Drosophila wilistoni. Keep that e-value in mind.
Now, instead of filtering the search results from the refseqRNA database, let’s create a custom search set. First, click Edit and Resubmit above the search summary.
Now, limit the search to animals entering animals in the Organism field. This parameter reduces the number of records in the search set.
Now, let’s look at the e-value from Drosophila wilistoni again.
It changed from 2e-47 to 1e-47, but all of the other values in the table stayed the same. it changes because the the e-value is dependent on the size of the search dataset. The larger the dataset, the more likely that you’ll get a coincidental match even though the sequences are related. Hence, the e-value went down as the size of the database went down.
I hope this tutorial has illustrated the importance of creating custom search sets and recording algorithm parameters.
Tobin Magle, Biomedical Sciences Research Support Specialist
Call for submissions – Art from the CU
Denver | Anschutz Medical Campus
Community 2016
The Exhibits Commi ee of the Health Sciences Library will be curating an exhibit of artwork created by faculty, staff and students of the University of Colorado Denver | Anschu Medical Campus. There are many talented artists at our two locations! This juried exhibition is an opportunity for us to learn about our talented co-workers, faculty, and students.
This exhibit will be on display November 4, 2015 — January 31, 2016 in the Gallery of the Health Sciences Library. An Opening Reception will be held on November 12, 2015 3:00-5:00 pm.
The Exhibits Commi ee is looking for submissions of all types of art created by
members of either CU Anschu or CU Denver! To submit artwork to be considered for inclusion in the show, please use the online form. The submission deadline is
September 25, 2015.
For more information, contact Debra Miller at debra.miller@ucdenver.edu or 303-724-2131.
Bioinformatics Bites: PubChem
This week’s bioinformatics bite is being outsourced to the NCBI Insights blog. Enjoy a concise explanation about how to identifying chemical targets to find cross reactions and prevent drug side effects:
h p://ncbiinsights.ncbi.nlm.nih.gov/2015/09/04/identifying-chemical-targets-finding-potential-cross-reactions-and-predicting-side-effects/
Have a nice long weekend,
Tobin Magle, PhD, Biomedical sciences research support specialist
Private: Online Service Interruption:
Labor Day, Monday, Sept 7, 6am-8am
The Office of Information Technology will be performing scheduled maintenance on Labor Day. The following services will be unavailablefrom 6:00am to 8:00 am
on Monday, September 7, 2015:
Off-Campus Access via PassportID to the library’s online resources: h ps://hsl-ezproxy.ucdenver.edu/login
(Access via employeeID/studentID with last name as password should still work.) UCDAccess (student and employee
portal): h ps://portal.prod.cu.edu/UCDAccessFedAuthLogin.html
Canvas (Learning Management System): h p://ucdenver.instructure.com/
MyAccount website (to claim your computer account): h ps://myaccount.ucdenver.edu
PassportID website (to change your password): h ps://passport.ucdenver.edu/
Sponsored User Request: h ps://passport.ucdenver.edu/
e-Directory (campus telephone directory): h ps://directory.ucdenver.edu/
Access to all other systems including email should NOT be affected during this maintenance.
If you encounter problems with the above services after the maintenance period has passed, please contact the OIT Help Desk:
303-724-HELP (303-724-4357)
h p://www.ucdenver.edu/its
Thank you in advance for your patience during this outage.
The Library Has Gone Green!
In the ongoing effort to become a more sustainable campus, the Health Sciences Library has installed WATER BOTTLE FILLING STATIONS. These machines encourage healthy habits and provide a convenient way for library users to refill their water bo les and encourage less use of plastic water bo les.
Currently there are three stations around the library. Stations are located on each floor by the restrooms on the south side of the building.
The stations are convenient, green, and wallet-friendly. Keep an eye out for new ones coming soon around campus.
Private: Test out the library’s new
responsive website
We’re happy to announce the soft launch of our newly designed responsive website — Test drive it today!
The new site will go live on Tuesday, September 8th. New features include:
Responsive design: The website is optimized for different screen sizes and devices,
including mobile. Adjust the width of your browser to see how the layout automatically changes.
Research Events: A calendar of campus-wide research events Exhibits: Featuring new exhibits at the library
Re-theme: The updated color pale e and theme of the CU Anschu website
Tell Us what you think — we look forward to your comments on our new website!
For questions, please contact: Vivienne Houghton, MLIS
Web Services Librarian & Senior Instructor 303-724-2178
vivienne.houghton@ucdenver.edu Thank you!
NCBI cracks 200 annotated
eukaryotic genomes
The National Center for Biotechnology Information (NCBI) is the central repository for molecular data in the US. They don’t generate their own data: all of the sequences in their databases is submi ed by external sources such as research labs, genomic sequencing consortia, and through the INSDC.
They do, however, provide an interface to access these data and create tools to analyze the data, which includes annotation a select set of genomes. Because NIH is primarily devoted to human health, they prioritize on eukaryotic genomes, especially mammals. The NCBI has annotated over 200 eukaryotic genomes so far. Do you want to see your favorite organism annotated by NCBI? They take requests through their Help Desk.
Clinical Corner: Inpatient
Glycemic Control
Today’s Internal Medicine Morbidity and Mortality Conference focused on a patient with type 1 diabetes who had difficulty with glycemic control during hospitalization. Here are several articles for background information:
Inpatient glycemic control: Best practice advice from the Clinical Guidelines Commi ee of the American College of Physicians. Qaseem A, Chou R, Humphrey
LL, Shekelle P; Clinical Guidelines Commi ee of the American College of
Physicians. Am J Med Qual. 2014 Mar-Apr;29(2):95-8. PMID: 23709472. Full text (free to UCD-AMC affiliates).
Management of hyperglycemia in hospitalized patients in non-critical care se ing: An Endocrine Society clinical practice guideline. Umpierrez GE, Hellman
R, Korytkowski MT, Kosiborod M, Maynard GA, Montori VM, Seley JJ, Van den Berghe G; Endocrine Society. J Clin Endocrinol Metab. 2012 Jan;97(1):16-38. PMID: 22223765. Full text (free to UCD-AMC affiliates).
Hospital management of hyperglycemia. Lleva RR, Inzucchi SE. Curr Opin Endocrinol Diabetes Obes. 2011 Apr;18(2):110-8. PMID: 21358407. Full text (free to UCD-AMC affiliates).
Intensive insulin therapy in hospitalized patients: A systematic
review. Kansagara D, Fu R, Freeman M, Wolf F, Helfand M. Ann Intern Med. 2011
Feb 15;154(4):268-82. PMID: 21320942. Full text (free to UCD-AMC affiliates). Three studies were presented at the end of conference as teaching points:
Patient communication during handovers between emergency medicine and internal medicine residents. Fischer M, Hemphill RR, Rimler E, Marshall S,
Brownfield E, Shayne P, Di Francesco L, Santen SA. J Grad Med Educ. 2012 Dec;4(4):533-7. PMID: 24294436. Free full text.
Exploring emergency physician-hospitalist handoff interactions: Development of the Handoff Communication Assessment. Apker J, Mallak LA, Applegate EB 3rd,
Gibson SC, Ham JJ, Johnson NA, Street RL Jr. Ann Emerg Med. 2010 Feb;55(2):161-70. PMID: 19944486. Full text (free to UCD-AMC affiliates).
Chart biopsy: An emerging medical practice enabled by electronic health records and its impacts on emergency department-inpatient admission handoffs. Hilligoss
B, Zheng K. J Am Med Inform Assoc. 2013 Mar-Apr;20(2):260-7. PMID: 22962194. Free full text.
Kristen DeSanto, MSLS, MS, RD, AHIP Clinical Librarian
Bioinformatics Bites: Primer BLAST
This week’s bioinformatics bites is going to look over the features of Primer BLAST. Back in my day (circa 2001-2014), we designed our primers by hand! Most of the places I worked, we’d have a printout of the genomic sequence we were working on that was annotated with transcripts and restriction site. We’d eyeball a good primer site and use OligoAnalyzer to find Tm and hairpins and self annealing and all that. One group I worked in would calculate Tm by counting up all the G’s and multiplying that by 4, then counting all the As and multiplying that by 2, and adding those two numbers together. There was no thought given to contaminating products in the design process. That was all trial and error in the PCR machine.
It was the dark ages.
Luckily, NCBI designed primer BLAST to help you all out. The primary function of this tool is primer design. First, enter a template.
(Click to enlarge images)
First, provide a unique identifier (accession or GI) for a record that’s in GenBank (like the NM_ number for an mRNA) or paste in a nucleotide sequence in FASTA format. i’m going to use the accession number (NM_001302688.1) for the human APOE gene mRNA.
Then, specify where you want your forward (sense) and reverse (antisense) primers to be in that sequence.
Finally, specify information about your primers (Tm) and desired PCR product
(length). You can also use this tool to create a matching primer for a preexisting primer.
We’ll use the example 5′-GGGAGCCCTATAATTGGACAAG-3′
If you used a refseq mRNA as your template, specify parameters involving introns
and exons, such as whether the primers span an exon-exon junction and how many
bases have to match on either side of the junction. You can also specify whether you want the product to span an intron on the genomic DNA.
Finally, specify how you want the algorithm to check for specificity of your primers by selecting your organism (or possible contaminating organism), the da a base you want to search, target size and specific primer parameters (mismatches etc.)
For this demo, I used the template NM_001302688.1 and specified that my forward primer is 5′-GGGAGCCCTATAATTGGACAAG-3′. I also specified that I want the product include on intron. All other values were kept as default. (Another handy feature is that the interface highlights non-default parameters in yellow.)
After clicking submit, a nice graphical summary of the results is displayed. Note that because i specified a forward primer, all the PCR products (in blue) start in the same place:
For each primer pair, the sequence of both primers, the product length, the size of the intron they span are included. along with information about Tm, length, start and stop positions, and self binding for each primer.
This is where the hack I mentioned last week comes in: because the primer that we entered into the search is also mapped onto the template, Primer BLAST effectively tells you the binding sure of the primer in terms of its position on the transcript we entered as the template. Pre y cool! Changing the template to the DNA accession number that contains this sequence would give us the genomic position of the primer, but we wouldn’t be able to do the cool stuff with introns.
Finally, it displays potential contaminating products you might see with human DNA as the PCR template:
In this case, it looks like we’d be amplifying another transcript variant of APOE. Definitely something to look out for when doing qPCR!
I hope you find this tool useful. Please let me know if I can help you with using primer BLAST or any other NCBI databases and tools.
Tobin Magle, Biomedical Sciences Research Support Specialist
New ProQuest Interface
On August 20th ProQuest will release an upgraded interface. The Health Sciences Library’s ProQuest resources include Dissertations & Theses.
Clinical Corner: Seizure/Epilepsy Videos
Cameron Ludt, DO (Neurology PGY-3) gave a presentation at Internal Medicine noon conference last Friday on epilepsy and seizure disorders. A highlight of the
presentation was the showing of several videos recorded on the epilepsy monitoring unit, so that students and residents could see firsthand what different types of seizures looked like.
To see even more seizure/epilepsy videos, the Health Sciences Library subscribes to the Neurology Video Textbook (2013), by Jonathan Howard from NYU School of Medicine. Here are the steps to browse and view videos:
Go to the following link: h p://online.statref.com/Notes/ResolveNote.aspx? NoteID=73052&grpalias=CU
Look for the table of contents on the lefthand column and click the arrow next to “Chapter 3. Seizures and Epilepsy” to open the menu:
Scroll down the page to view all available videos.
Please contact me for further questions or if you have problems accessing the book. Kristen DeSanto, MSLS, MS, RD, AHIP
Clinical Librarian
kristen.desanto@ucdenver.edu • 303-724-2121
Bioinformatics bites: How do I find
primer binding sites?
This week’s bioinformatics bite comes from another actual patron question (paraphrased):
I have all these primers that someone else designed. How to I figure out where they bind and what they amplify?
Disclaimer: this isn’t actually the answer I gave to the person seeking help, but I’ve since found a more efficient tool.
Probably the fastest way to get this information is to use a simple tool called Primer Map.
Map these Primers: (reverse) aacagctatgaccatg, (T3) a aaccctcactaaag, (KS) cgaggtcgacggtatcg, (SK) tctagaactagtggatc, (T7) aatacgactcactatag, (-40) g cccagtcacgac, (Sp6) a taggtgacactatag, (M13 for) gtaaaacgacggccagt, (M13 rev) cacacaggaaacagctatgaccat, (BGH rev) tagaaggcacagtcgagg, (pGEX for) ctggcaagccacg tggtg, (pGEX rev) ggagctgcatgtgtcagagg, (T7-EEV aaggctagagtac aatacga, (pUC/M13 Forward) g cccagtcacgac, (pUC/M13 forward) cgccaggg cccagtcacgac, (pUC/M13 reverse) caggaaacagctatgac, (pUC/M13 reverse) tcacacaggaaacagctatgac, (Glprimer1) tgtatc atggtactgtaactg, (GLprimer2) c tatg tggcgtc cca, (RVprimer3) ctagcaaaataggctgtccc, (RVprimer4) gacgatagtcatgccccgcg,
(Lambda gt11 Forward) ggtggcgacgactcctggagcccg, (Lambda gt11 Reverse) gacaccagaccaactggtaatg, (Lambda gt10 Forward) c gagcaag cagcctgg aag, (lambda gt10 Reverse) gaggtggc atgagta tc ccagggta, (Pinpoint Sequencing) cgtgacgcggtgcagggcg,
(pTarget Sequencing) acgccaag a taggtgaca
To this sequence:
>sample sequence
cagctggggggaggtggcgaggaagatgacgtggtcgaggtcgacggtatcgag gtcgcggcagctgccaatacgactcactatagaggagaagtagcaagaaaaataacatgataa atcacgacaactacctggtgatg
Once the template is in the top box, and the primers are in the bo om box, hit Submit. (The output gets a lot cleaner if you turn translation and restriction enzyme displays off in the se ings.)
The results pop up in a new window. The first results section show where the primers bind, with forward (sense) primers highlighted in purple and reverse (antisense) primers highlighted in orange.
The second part of the results page show a table with all the primers that you input, highlighting which ones that bound with the color that indicates their orientation:
The only thing that is missing is a their column that indicates the position on the template at which the primers bind. I guess beggars can’t be choosers though. Next time I’ll show you how to do this using NCBI’s Primer BLAST. This algorithm is actually build for primer design, but it can be hacked for this purpose and provides be er visualizations and more information.
Tobin Magle, PhD, Biomedical Sciences Research Support Specialist
Private: Off-Campus Access
is changing!
On August 26, the Health Sciences Library will be making changes to our off-campus proxy system so that you can now log in with your PassportID (this is
the same username and password you use for other University resources – like o365 email and the UCDAccess Portal).
Why are we making this change?
Login via PassportID is more secure for you, the user. It will also be far easier for the library to manage. Just one example: in the past there has been a slight delay of up to a week before new students and employees were able to have access to off campus resources. That delay will now disappear.
Here is a screenshot of the new login page that will be in place starting 8 am, Aug. 26 2015 (click to enlarge):
There will be some exceptions:
University of Colorado Hospital employees will still need to log in the old way,
using their username of: le er H followed by their UCH employee ID number (eg: Hxxxxxx). Password is user’s last name.
Volunteer Clinical Faculty (VCF program) will continue to log in with their
assigned ID number as user name and last name as password.
As this is a “soft launch” , the old way of logging in will still work for a limited time only: user name as employee ID number, and last name as password.
However, if you had a web browser automatically enter your login and password,
you will need to clear out the saved password. If you have any trouble with this,
please call our library Service Desk for directions on clearing saved password from various browsers.
Difficulty logging in? Please contact the HSL Service Desk at 303-724-2152 or use Ask
Us.
Questions? Concerns? Please contact Jeff.Kun man@ucdenver.edu
New article linker enhancement:
Sidebar Helper
Tell us what you think about the new article linker enhancement: Sidebar Helper
frame. Sidebar Helper will help you connect to full text and stay connected to library
services in the places where you are doing research, such as journal websites and PubMed.
We are testing Sidebar Helper from August 11th, 2015 to August 21st, 2015. We appreciate your feedback!
To see Sidebar Helper in action, go to the library’s home page and do a
search in PubMed, Google Scholar, or other journal sites where you are used to seeing the Article Linker bu on.
Bioinformatics Bites: R tutorials
for beginners
Today’s Bioinformatics Bite is based on a hypothetical question that I think a lot of people are afraid to ask:
I hear R is a great tool for doing bioinformatic analysis, but I have no idea how to code. How can I get started?
Well, I’d say the first step is to Install R.
Windows Mac OSX
Linux (though if you’re using Linux, you probably don’t need this tutorial) This first installation installs the R coding language and a bare-bones editor to write and run code in.
If you want a nicer interface, I’d suggest installing R studio. which has a lot of bells and whistles that will made using R a lot easier. R Studio contains a text editor with
highlighting, integrated help functions, an environment window that reminds you what variables you created and a console that allows you to execute your code right from the editor.
But how do you even know where to begin now that you have it installed? A good place to start for R basics is Swirl. Swirl is a tutorial system that you can use INSIDE R. All you have to do to install Swirl is type
inside the R console, and swirl will be automatically installed! Now to run Swirl, just type
> library("swirl")
> swirl()
in the R console to load the swirl library and run the tutorials. Then you can pick from a variety of introductory tutorials that are closely linked to courses in the Johns
Hopkins University Data Science Specialization on Coursera. Now we just need to get someone to write some Swirl tutorials for Bioconductor.
If you have any questions about how to set up R, don’t hesitate to Ask. Tobin Magle,
Biomedical Sciences Research Support Specialist tobin.magle@ucdenver.edu
Clinical Corner: High-Value,
Cost-Conscious Care
Steven Weinberger MD, of the American College of Physicians (ACP), gave a
presentation on Friday, July 31 at internal medicine conference on the topic of high-value, cost-conscious care. The ACP High Value Care initiative has two aims:
1. Helping physicians to provide the best possible patient care.
2. Simultaneously reducing unnecessary costs to the healthcare system. Here are resources to learn more about this topic:
Choosing Wisely®
www.choosingwisely.org
Choosing Wisely® is an initiative of the ABIM (American Board of Internal Medicine) Foundation, which explores ways to avoid unnecessary medical interventions. The website contains recommendations from over 70 medical specialty societies and health care organizations.
ACP High Value Care
hvc.acponline.org
The ACP High Value Care site contains guidelines and clinical recommendations that address the benefits, harms, and costs of various interventions. There are also
curriculum and public policy recommendations, and resources for consumers.
Ann Intern Med. 2013 Jan 1;158(1):55-9.
Design and use of performance measures to decrease low-value services and achieve cost-conscious care.
Baker DW, Qaseem A, Reynolds PP, Gardner LA, Schneider EC; American College of Physicians Performance Measurement Commi ee.
Abstract
Improving quality of care while decreasing the cost of health care is a national priority. The American College of Physicians recently launched its High-Value Care Initiative to help physicians and patients understand the benefits, harms, and costs of interventions and to determine whether services provide good value. Public and private payers continue to measure underuse of high-value services(for example, preventive services, medications for chronic disease),but they are now widely using performance measures to assess use of low-value interventions (such as imaging for patients with
uncomplicated low back pain) and using the results for public reporting and pay-for-performance. This paper gives an overview of performance measures that target low-value services to help physicians understand the strengths and limitations of these measures, provides specific examples of measures that assess use of low-value services, and discusses how these measures can be used in clinical practice and policy.
Ann Intern Med. 2012 Aug 21;157(4):284-6.
Teaching high-value, cost-conscious care to residents: the Alliance for Academic Internal Medicine–American College of Physicians Curriculum.
Smith CD; Alliance for Academic Internal Medicine–American College of Physicians High Value; Cost-Conscious Care Curriculum Development Commi ee.
Abstract
Health care expenditures are projected to reach nearly 20% of the U.S. gross domestic product by 2020. Up to $765 billion of this spending has been identified as potentially avoidable; many of the avoidable costs have been a ributed to unnecessary services. Postgraduate trainees have historically received li le specific training in the
stewardship of health care resources and minimal feedback on resource utilization and its effect on the cost of care. This article describes a new curriculum that was developed collaboratively by the Alliance for Academic Internal Medicine and the American College of Physicians to address this training gap. The curriculum introduces a simple, stepwise framework for delivering high-value care and focuses on teaching trainees to incorporate high-value, cost-conscious care principles into their clinical practice. It consists of ten 1-hour, case-based, interactive sessions designed to be flexibly
incorporated into the existing conference structure of a residency training program.
Ann Intern Med. 2012 Jan 17;156(2):147-9.
Appropriate use of screening and diagnostic tests to foster high-value, cost-conscious care.
Qaseem A, Alguire P, Dallas P, Feinberg LE, Fi gerald FT, Horwitch C, Humphrey L, LeBlond R, Moyer D, Wiese JG, Weinberger S.
Abstract
Unsustainable rising health care costs in the United States have made reducing costs while maintaining high-quality health care a national priority. The overuse of some screening and diagnostic tests is an important component of unnecessary health care costs. More judicious use of such tests will improve quality and reflect responsible awareness of costs. Efforts to control expenditures should focus not only on benefits, harms, and costs but on the value of diagnostic tests-meaning an assessment of whether a test provides health benefits that are worth its costs or harms. To begin to identify ways that practicing clinicians can contribute to the delivery of high-value, cost-conscious health care, the American College of Physicians convened a workgroup of physicians to identify, using a consensus-based process, common clinical situations in which screening and diagnostic tests are used in ways that do not reflect high-value care. The intent of this exercise is to promote thoughtful discussions about these tests and other health care interventions to promote high-value, cost-conscious care.
Ann Intern Med. 2011 Sep 20;155(6):386-8.
Providing high-value, cost-conscious care: a critical seventh general competency for physicians.
Weinberger SE.